More Hints for the Plugin Author
The diffKK plugin (at
plugins/xafs/diffkk.py)
implements the differential Kramers-Kronig transform
suggested by Cross and Newville
using the MacLaurin series algorithm suggested by Ohta and Ishida,
Applied Spectroscopy 42:6 (1988) pp. 952-957. This is a way of
matching experimental data to tabulated values of the imaginary part
of the energy-dependent correction to the Thompson scattering factor
and using a
Kramers-Kronig transform
to generate the real part. The advantage of this over the tabulated
values is that, starting from a measured XAS spectrum, the diffkk
results include the effect of the local scattering environment on the
energy-dependent scattering function.
The diffKK plugin uses the MBACK plugin to do the matching of the XAS data to tabulated f′′(E) data. It also demonstrates a few more features of Larch plugins that merit discussion in this tutorial.
A Larch plugin implementing a Group
The MBACK plugin implements one function, which is registered to Larch's symbol table. This function is intended for use with an existing data Group.
The diffKK plugin takes a slightly different approach. It too
exposes a function in Larch's symbol table. This function is called
diffkk and returns a reference to a diffKK Group. Here's what
that looks like:
def diffkk(energy=None, mu=None, z=None, edge='K', mback_kws=None, _larch=None, **kws):
"""
Make a diffKK group given mu(E) data
Attributes
energy: energy array
mu: array with mu(E) data
z: Z number of absorber
edge: absorption edge, usually 'K' or 'L3'
mback_kws: arguments for the mback algorithm
"""
return diffKKGroup(energy=energy, mu=mu, z=z, mback_kws=mback_kws, _larch=_larch)
def registerLarchPlugin(): # must have a function with this name!
return ('_xafs', { 'diffkk': diffkk })
and here how it is called:
:FIXME: angle bracket wierdness
larch> dkk = diffkk(data.energy, data.xmu, z=29, edge='K', mback_kws={'e0':8979, 'order':4})
larch> show c
== diffKK Group: 7 symbols ==
edge: 'K'
energy: array<shape=(612,), type=dtype('float64')>
kk: <bound method diffKKGroup.kk of <diffKK Group>>
mback_kws: {'e0': 8979, 'order': 4}
mu: array<shape=(612,), type=dtype('float64')>
plotkk: <bound method diffKKGroup.plotkk of <diffKK Group>>
z: 29
The diffkk function creates and returns a diffKK Group, which is
called dkk. This Group contains the energy and μ(E) arrays,
the scalar values of z and edge arguments, the dictionary value of
the mback_kws argument, and references to a couple of functions,
kk() and plotkk(). This really underscores the definition of a
Group being a collection of things.
The diffKK Group is defined in the plugin as a class which inherits from Larch's Group. That is, diffKK has all the properties of a Larch Group and the additional properties given to it in this plugin.
class diffKKGroup(Group):
"""
A Larch Group for generating f'(E) and f"(E) from a XAS measurement of mu(E).
"""
def __init__(self, energy=None, mu=None, z=None, edge='K', mback_kws=None, _larch=None, **kws):
kwargs = dict(name='diffKK')
kwargs.update(kws)
Group.__init__(self, **kwargs)
self.energy = energy
self.mu = mu
self.z = z
self.edge = edge
self.mback_kws = mback_kws
if _larch == None:
self._larch = Interpreter()
else:
self._larch = _larch
def __repr__(self):
return '<diffKK Group>'
In typical fashion, the class has a document string. The __init__
method of the class is defined and used to initialize various
attributes of the class. Most of these attributes are the things that
will be needed to perform the differential KK transform. It also
defines a _larch reference which will point at the correct value of
the Larch interpreter.
The __repr__ method is also specified so that the class has an
unambiguous string representation:
larch> print dkk
<diffKK Group>
The plugin then goes on to define the kk() and plotkk() functions.
Using the dkk group defined by the commmand above, you can perform
the KK transform and plot the results. These commands post a plot
to the screen:
larch> dkk.kk()
larch> dkk.plotkk()
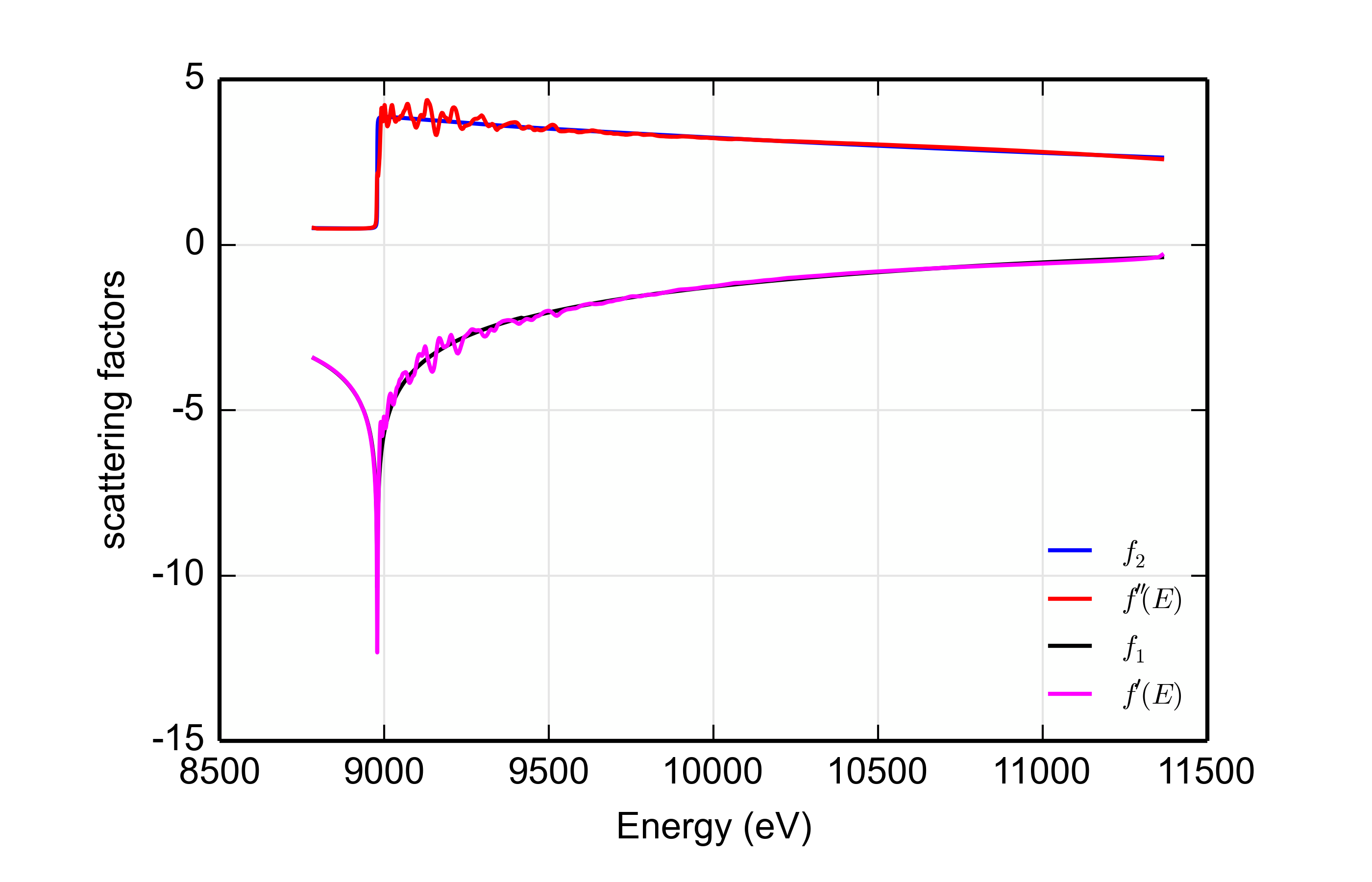
The kk() function places additional arrays and scalars in the dkk
Group.
This kind of plugin -- one which defines a Group and stores a tightly connected, curated collection of data and function references -- is very handy. It makes a nice, neat package for an analysis concept. See the section on Feff paths from the Larch document for several more examples of this sort of plugin.
Using NumPy effectively
The diffKK plugin also makes effective use of NumPy's vectorized calculations. At the heart of the diffKK algorithm is a computationally intensive KK transform. Here is a scalar implementation of the MacLaurin series algorithm
def kkmclr_sca(e, finp):
"""
reverse (f''->f') kk transform, using maclaurin series algorithm
"""
npts = len(e)
fout = [0.0]*npts
factor = -FOPI * (e[npts-1] - e[0]) / (npts - 1)
nptsk = npts / 2
for i in range(npts):
fout[i] = 0.0
ei2 = e[i]*e[i]
ioff = i%2 - 1
for k in range(nptsk):
j = k + k + ioff
de2 = e[j]*e[j] - ei2
if abs(de2) <= TINY:
de2 = TINY
fout[i] = fout[i] + e[j]*finp[j]/de2
fout[i] *= factor
return fout
(I have removed most of the document string and all of the exception handling for the sake of brevity.)
This has nested loops over the entire range of the arrays containing the data to be transformed. While the inner loops is smaller by a factor of 2 than the outer loop, this is effectively of order
(n2) in calculation time. It scales poorly in time as the arrays become large.
Here is how this is implemented in diffKK:
def kkmclr(e, finp):
"""
reverse (f''->f') kk transform, using maclaurin series algorithm
"""
npts = len(e)
fout = np.zeros(npts)
factor = -FOPI * (e[-1] - e[0]) / (npts-1)
ei2 = e**2
ioff = np.mod(np.arange(npts), 2) - 1
nptsk = npts/2
k = np.arange(nptsk)
for i in range(npts):
j = 2*k + ioff[i]
de2 = e[j]**2 - ei2[i]
fout[i] = sum(e[j]*finp[j]/de2)
fout = fout * factor
return fout
Here NumPy's vectorization is used to remove the inner loop. Calculations are made on entire vectors rather than by iterating over the elements of the inner loop. The iteration still happens, but it is done behind the scenes using NumPy's efficient vectorization. This is not quite as fast an order n calculation, but it is much faster than the scalar implementation above. On the modest and fairly old computer I am sitting at to write this, the scalar calculation takes about 15 seconds while the vectorized calculation takes under 2 seconds.
Take the time to understand how this vectorized math stuff works. It pays off!